Using recurring spatially contiguous substructures in the Protein Database for protein structure prediction
Abstract
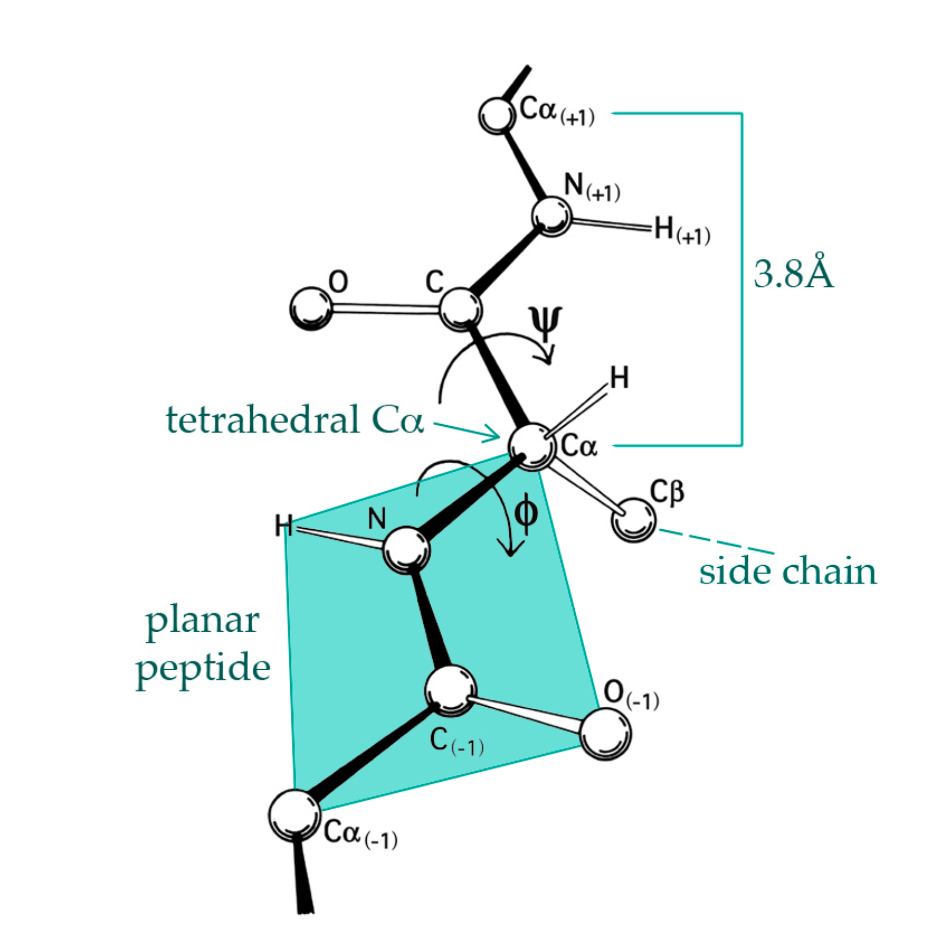
Since we knew that the protein’s sequence encodes its structure, researcher have been trying to predict the structures computationally. Challenged by the vastness of the conformational space, researchers have leveraged the similarities present in the Protein Data Base to guide prediction. In this work, we present a new approach for protein structure prediction using a novel source of information: Building Blocks. These are sequentially non-continuous, but structurally contiguous, structural motifs that are retrieved according to a given sequence.
We devised two approaches that uses Building Blocks to improve the prediction and compared them. The first, the foldtree approach, construct structures by sampling the Building Blocks all through the search, while the second, the constraint approach, alter the energy landscape to guide the search towards structures obeying the Building Blocks’ spatial arrangement. The two algorithms were able to significantly improve the prediction compared to the uninformed method. The constraint approach showed better performance than the foldtree approach, although it suffered from long run times and did not tolerate erroneous input.
We have shown that a new source of information can be successfully leveraged for ab initio structure prediction. This work lays out the basis for future research on utilizing this information and ultimately making a significant contribution towards the solution of the structure prediction problem.